Data management plan for liquid and vapour isotope measurements
This data management plan comprises the generation, processing and storage of data files from liquid and vapour isotope measurements with Picarro instruments at FARLAB.
1. Data flow for Liquid water isotope measurements
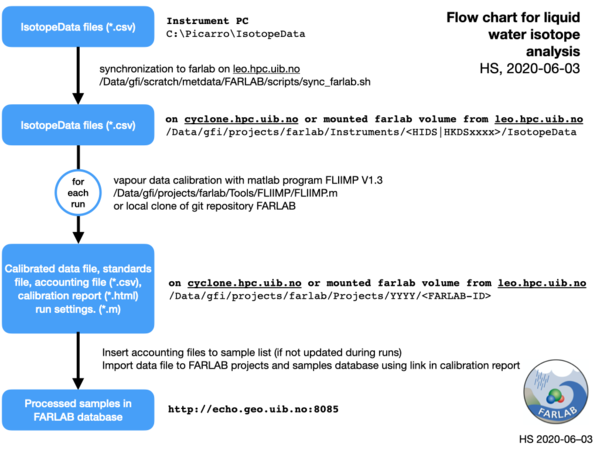
- Each project registered in the FARLAB projects data base will have the associated processed files located in a subfolder on the FARLAB volume according to the naming scheme
farlab/Projects/<YYYY>/<YYYY-ID-PI>where YYYY is the year the project was registered, ID is the FARLAB project ID assigned by the FARLAB project data base, and PI is a two-letter code indicating the handling FARLAB member. - Injection result files (*.csv format) are generated on the instrument PC and stored in folder C:\Picarro\IsotopeData
- Injection result files are synced routinely every ca. 10 min to a backup volume from LAB-IT (
felles2.uib.no:/LABIT1/labit_geo_farlab). A script routinely synchonizes the data files to the farlab volume, mounted onleo.hpc.uib.no:/Data/gfi/projects/farlab/Instruments/<serial_number>/IsotopeData. - For each run, liquid injection data have to be calibrated manually with the matlab program FLIIMP. FLIIMP (currently in version 1.3) is shared on the FARLAB UiB git repository.
- All the data files resulting from FLIIMP calibration, contained in a folder created by FLIIMP, and including the accouting, standards, results, and all file (*.csv format), the calibration report (*.html format), images (*.png) and settings files for each run (*.mat) should be located on the FARLAB volume in the respective projects folder.
- The processed samples are imported to the FARLAB database either manually using the Samples and Results import link, or with the corresponding upload link in the FLIIMP (forthcoming V1.4).
Data flow for Water vapour isotope measurements
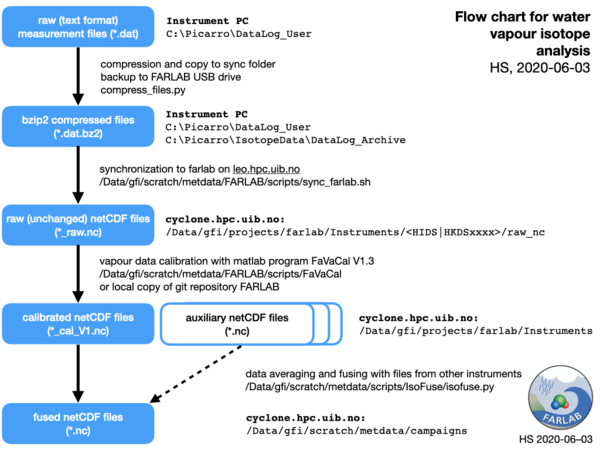
- Each project registered in the FARLAB projects data base will have the associated processed files located in a subfolder on the FARLAB volume according to the naming scheme
farlab/Projects/<YYYY>/<YYYY-ID-PI>where YYYY is the year the project was registered, ID is the FARLAB project ID assigned by the FARLAB project data base, and PI is a two-letter code indicating the handling FARLAB member. - Vapour isotope result files (*.dat format, text-based) are generated on the instrument PC and stored in folder C:\Picarro\DataLog_User.
- A compression routine (compress_dat.py) continuously bzip2-compresses the hourly *.dat files (*.dat.bz2 files) and copies them to the subfolder
C:\Picarro\IsotopeData\DataLog_Archive. In addition, the contents of directory DataLog_User is continuously copied to the backup drive, if attached. - Compressed dat files are synced routinely every ca. 10 min to a backup volume from LAB-IT (
felles2.uib.no:/LABIT1/labit_geo_farlab). A script routinely synchonizes the data files to Instrument subfolderds on the farlab volume, mounted onleo.hpc.uib.no:/Data/gfi/projects/farlab/Instruments/<serial_number>/DataLog_User. - Text-based, compressed data files (*.dat.bz2) are converted to nc4 compressed netCDF format without correction or calibration (raw netCDF files, *_raw.nc). Conversion runs automatically from script
/Data/gfi/scratch/metdata/scripts/FARLAB/convert_FARLAB_raw_daily.shas cron job, which call the generic conversion routine convert_L21xxi2nc.sh. - Raw netCDF files are calibrated and processed (corrected, averaged) with the matlab software FaVaCal (current version 1.3). Processing is commonly done for up to 1-2 months in a row. There are different options to provide calibration information, from SDM use, manual injections, or as pre-specified file, as described in the user manual.
- Calibrated netCDF data files (*.nc) and the calibrations file (*.csv) should be located in the FARLAB project directory, and can from there be shared with clients or used in further processing and analysis.
- An optional further step is to fuse data files with the software utility IsoFuse. IsoFuse provides joint, averaged netCDF data files based on any CF-compatible input data files. In a two stage process, the files to be fused are first scanned for variables and time resolution. In a second step, the files are fused to a common time scale, with optional application of time shifts and scaling/offset. See the metdata campaigns and scripts folder for examples.